Beyond NGS: Bionano Saphyr Next Generation Mapping
By Catherine Brownstein, MPH, PhD
Assistant Director, Molecular Genetis Core Facility
May 25, 2022
Large and small scale structural variants in the human genome might modulate gene expression in ways that influences physiological response and/or the clinical course of disease. Many of these structural variants go unrecognized using standard Next Generation Sequencing (NGS).
Limitations of Next Generation Sequencing
Next Generation Sequencing studies often conclude without an explanation for a patient’s condition. This may be due to technical limitations. Structurally complex loci underlie many disorders which can be challenging to resolve using currently available methods such as: karyotyping, clinical array, PCR-based tests, and NGS alone. On average, exome and genome sequencing solves only 25-33% of cases. Two-thirds of the human genome consists of repetitive sequences, and the short-read sequences produced by traditional exome and genome map with poor accuracy to these repeats. In addition, alignment algorithms typically fail to identify the exact genomic location for short-reads. When they do align, the limited 100-150 bp read length and spacing of paired-end reads does not allow for a correct sizing of larger repeats. NGS also misses most structural variation (SV) such as balanced translocations and inversions, both of which can result in disease.
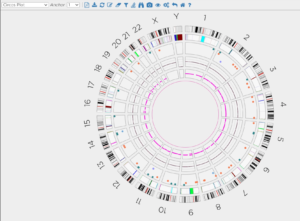
Dynamic Circos
Bionano Genomics Saphyr System
The Bionano Genomics Saphyr System overcomes these limitations. It utilizes a high-throughput, genome-wide “optical mapping” technology to interrogate structural differences in the genome and increase the contiguity and accuracy needed to produce quality genome assemblies. Structural variants and repeats are measured directly within long, single-molecule “reads” for comprehensive analysis of what has been dubbed “the inaccessible genome.” Bionano maps are built without any reference guidance or bias. This differentiates Optical Genome Mapping (OGM) from NGS, where short-read sequences are typically aligned to a reference. Only de novo constructed genomes, like Bionano maps, allow for a completely unbiased, accurate assembly. Bionano’s
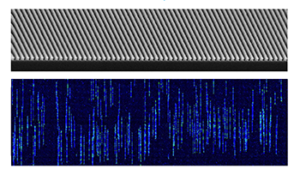
Linearized DNA moving through a nanochannel array
SVs are observed, not inferred. When short-read NGS sequences are aligned to the reference genome, algorithms piece together sequence fragments in an attempt to rebuild the actual structure of the genome. With OGM, megabase-size native DNA molecules are imaged, and large SVs or their breakpoints (in the case of inter-chromosomal translocations) can be observed directly in the label pattern on the molecules. In a published study, the Bionano System was able to identify 909 insertions and 661 deletions (including 800 novel insertions) that were previously unidentified in the 1000 Genomes Project (1).
Bionano Saphyr and the IDDRC MGCF during COVID-19
When COVID-19 hit, the scientific community, with the support of Bionano Genomics, quickly formed a consortium to perform Bionano Saphyr OGM on patients with a severe course of COVID-19, called the Host Genome Structural Variant Research Consortium [https://www.covid19hostgenomesv.org/]. The IDDRC Molecular Genetics Core Facility was able to contribute and this work has culminated in one publication (2), to date. The OGM approach was complimentary to other Next Generation Sequencing projects ongoing at BCH and across the country.
Bionano Saphyr at the IDDRC MGCF
The Molecular Genetics Core Facility of our IDDRC has unparalleled expertise on the Bionano Saphyr and one of the only OGM systems in Boston. IDDRC users have access to our advice on study design and preparation. We can also price out larger projects as price can decrease with a guaranteed number of samples for bulk purchasing. We are here to help!
Visit us today:
Gene Analysis Group | Molecular Genetics Core Facility
Intellectual and Developmental Disabilities Center
Boston Children’s Hospital
Email Us
(1) Mak ACY, Lai YYY, Lam ET, Kwok T-P, Leung AKY, Poon A, Mostovoy Y, Hastie AR, Stedman W, Anantharaman T, Andrews W, Zhou X, Pang AWC, Dai H, Chu C, Lin C, Wu JJK, Li CML, Li J-W, Yim AKY, Chan S, Sibert J, Džakula Ž, Cao H, Yiu S-M, Chan T-F, Yip KY, Xiao M, Kwok P-Y. Genome-Wide Structural Variation Detection by Genome Mapping on Nanochannel Arrays. Genetics. 2016;202(1):351-62. Epub 2015/10/28. doi: 10.1534/genetics.115.183483. PubMed PMID: 26510793.
(2) Sahajpal NS, Jill Lai CY, Hastie A, Mondal AK, Dehkordi SR, van der Made CI, Fedrigo O, Al-Ajli F, Jalnapurkar S, Byrska-Bishop M, Kanagal-Shamanna R, Levy B, Schieck M, Illig T, Bacanu SA, Chou JS, Randolph AG, Rojiani AM, Zody MC, Brownstein CA, Beggs AH, Bafna V, Jarvis ED, Hoischen A, Chaubey A, Kolhe R. Optical genome mapping identifies rare structural variations as predisposition factors associated with severe COVID-19. iScience. 2022;25(2):103760. Epub 2022/01/18. doi: 10.1016/j.isci.2022.103760. PubMed PMID: 35036860; PMCID: PMC8744399.