Editing the mouse and human genomes to reveal the formation, function, and diseases of the nervous system, and help identify new therapies
Employing IDDRC Cores for complex, state of the art genetic research and screening
By Clifford Woolf, MB, BCh, PhD
Director, F.M. Kirby Neurobiology Center, Boston Children’s Hospital
Co-Core Director, Genetic Analysis and Editing Core, IDDRC
Professor of Neurobiology and Neurology, Harvard Medical School
The successful sequencing of the 3 billion base pairs of the human genome by the Human Genome Project revealed ~30,000 genes, which at least for me, was surprisingly few, given the enormous complexity of human biology and the very broad diversity of inherited human disease. This is particularly true for the nervous system, which comprises billions of neurons and trillions of synaptic connections engineered in a tightly organized circuit format. How do relatively so few genes control the development of the highly complex nervous system and ensure its healthy diverse functions, and which genes are drivers of neurological/psychiatric disease or disease risk?
Introduction of CRISPR/Cas9 Gene Editing
For a long while it was difficult to think of ways of addressing this in a comprehensive way, other than the slow and expensive knockout of single candidate genes and transgenic gene overexpression in rodents and other preclinical model organisms. However, the introduction of CRISPR/Cas9 gene editing in the last decade suddenly completely changed this. We can now target every gene in multiple ways, reducing or abolishing activity (CRISPR inhibition), explore the effects of variants/mutations, test the consequences of increasing transcription (CRISPR activation), and use reporters to monitor expression dynamically or to manipulate the cells that express the gene. In some cases, we can now conduct unbiased genome-wide gain and loss of function screens.
CRISPR/Cas9 utilizes target-specific guide RNA, Cas9 protein and a donor template introduced by microinjection into one-cell embryos or stem cells and relies on the combined effect of target sequence specific guide RNA and Cas9 nuclease induced double strand break. Cellular mechanisms repair the double strand breaks by two pathways, either non-homologous end joining, or homology directed repair. Non-homologous end joining is an error prone but highly efficient mechanism that actively repairs double strand breaks and introduces insertion or deletion of nucleotides, often resulting in a premature stop codon, leading to loss of gene function, creating a gene knockout. Homology directed repair is a less efficient but more precise repair mechanism and is used to introduce accurate modifications by supplying a single or double strand DNA donor template with specific changes, flanked by homology arms.
Targeting the genetic instructions of the nervous system
Gene-editing and gene-manipulation are total game changers which are helping us to begin to comprehensively address the extraordinary complexity of the nervous system by enabling us to target the genetic instructions of the system. I currently use gene editing in several ways.
Gene Editing Core: Assessing functionality of genes in mice
First, working with Mantu Bhaumik and his team in the IDDRC Gene-Editing Core, I manipulate those genes in the mouse that I suspect from single cell transcript profiling studies (Chiu et al, 2014; Renthal et al, 2020), and human genetics (Tegeder et al, 2006), to be involved in different aspects of nervous system function, determining the effects of altering their transcription, using them to drive expression of technological tools for optogenetic or chemogenetic modifications, or even ablating, defined sets of cells to assess their function (Chiu et al 2013; Browne 2017; Michoud et al 2021). While in some cases this reveals the function of novel pathways in defined pathological conditions – like neuropathic pain (Latremoliere et al, 2015) or the regenerative response to nerve injury (Cheng et al, 2021), in others it reveals how the mouse cannot be a simple surrogate for particular single gene driven human diseases; single gain-of function mutations in the voltage-gated sodium channel Nav1.7 produce severe spontaneous pain in patients but no detectable pain-like behavior in mice (Chen et al, 2021)!
Our phenotyping of the engineered mice involves use of multiple IDDRC cores for sequencing, cell profiling, behavioral and physiological measurements, and imaging. A major focus of the lab is now recording from neurons in the spinal cord and cortex and identifying their role in various aspects of pain, and this is made possible by being able to target GCaMP in defined neuronal populations or the use of designer receptors exclusively activated by designer drugs (DREADS) to temporarily suppress or activate defined neuronal populations (Liu et al, 2018).
Human Neuro Core: Investigating Human Cell Biology and Diseases in iPSC derived neurons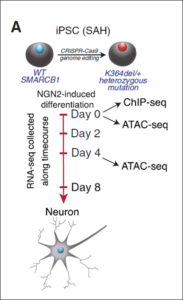
My second major exploitation of gene editing is in the context of human stem cell biology, working closely with the IDDRC Human Neuron Core. Gene editing enables the generation of isogenic controls for familial diseases such as ALS, by correcting the mutation in a patient’s iPSC line (Kiskinnis et al, 2014), enabling disease mechanisms to be identified (Wainger et al, 2014) and screens to be run to identify disease targets (Huang et al, 2021). A particular exciting aspect of this work is that the reduction of the hyperexcitability produced in familial ALS patient iPSC derived motor neurons by the Kv7 potassium channel opener retigabine in a dish (Wainger et al, 2014) was fully predictive of similar effects in patients in vivo (Wainger et al, 2021), showing how stem cell disease models can be used to identify drugs that convert diseased cells to healthy ones, and that this can lead to novel therapeutics (Boivin et al, 2022).
A second stem cell modeling effort of ours is for pain. In a major collaboration with NCATS and HMS, we have developed multiple human neuronal phenotypic screens designed to enable selective silencers of nociceptors to be screened for and new interventions to reduce inflammatory and neuropathic pain to be discovered (Jayakar et al, 2021).
Finally, in a collaboration with Cigall Kadoch of DFCI, we have been exploring how mammalian switch/sucrose non-fermentable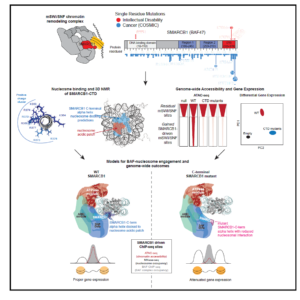 (mSWI/SNF) complexes act as multi-component machines to remodel chromatin architecture (Valencia et al, 2019). We started off by examining the molecular, structural, and genome-wide regulatory consequences of recurrent, single-residue mutations in the putative coiled-coil C-terminal domain of the SMARCB1 (BAF47) subunit, which cause the intellectual disability disorder Coffin-Siris syndrome (CSS), and which are also found in some cancers. The SMARCB1 C-terminal domain contains a basic α helix that binds directly to the nucleosome acidic patch and all Coffin-Siris syndrome-associated mutations disrupt this binding. Furthermore, these mutations abrogate mSWI/SNF-mediated nucleosome remodeling activity and enhancer DNA accessibility, without any changes in genome-wide complex localization. Heterozygous Coffin-Siris syndrome-associated SMARCB1 mutations result in dominant gene regulatory and morphologic changes during iPSC-neuronal differentiation. These gene editing-based stem cell studies have unmasked an evolutionarily conserved structural role for the SMARCB1 C-terminal domain that is perturbed in human neurodevelopmental disease, and we are now conducting further studies on additional SMARCB1 gene-edited cell lines.
(mSWI/SNF) complexes act as multi-component machines to remodel chromatin architecture (Valencia et al, 2019). We started off by examining the molecular, structural, and genome-wide regulatory consequences of recurrent, single-residue mutations in the putative coiled-coil C-terminal domain of the SMARCB1 (BAF47) subunit, which cause the intellectual disability disorder Coffin-Siris syndrome (CSS), and which are also found in some cancers. The SMARCB1 C-terminal domain contains a basic α helix that binds directly to the nucleosome acidic patch and all Coffin-Siris syndrome-associated mutations disrupt this binding. Furthermore, these mutations abrogate mSWI/SNF-mediated nucleosome remodeling activity and enhancer DNA accessibility, without any changes in genome-wide complex localization. Heterozygous Coffin-Siris syndrome-associated SMARCB1 mutations result in dominant gene regulatory and morphologic changes during iPSC-neuronal differentiation. These gene editing-based stem cell studies have unmasked an evolutionarily conserved structural role for the SMARCB1 C-terminal domain that is perturbed in human neurodevelopmental disease, and we are now conducting further studies on additional SMARCB1 gene-edited cell lines.
All this human cell work is heavily dependent on the Assay Development and Screening Facility (ADSF) of the Human Neuron Core, run by Lee Barrett.
In sum then, gene editing provides exciting multiple diverse opportunities to study the nervous system, and the IDDRC Cores at BCH provide the necessary platform to enable this.
*******
Bibliography
Browne LE, Latremoliere A, Lehnert BP, Grantham A, Ward C, Alexandre C, Costigan M, Michoud F, Roberson DP, Ginty DD, Woolf CJ. Time-Resolved Fast Mammalian Behavior Reveals the Complexity of Protective Pain Responses. Cell Rep. 2017 Jul 5;20(1):89-98.
Boivin B, Roet K, Huang X, Karhohs K, Rohban M, Sandoe J, Wiskow O,Maeda R, Grantham A, Dornon M, Shao J, Frost D, Baker D, Eggan K, Carpenter A, Woolf CJ. A Multiparametric Activity Profiling Platform for Neuron Disease Phenotyping and Drug Screening. Molecular Biology of the Cell (2022) In Press.
Chen L, Wimalasena NK, Shim J, Han C, Lee SI, Gonzalez-Cano R, Estacion M, Faber CG, Lauria G, Dib-Hajj SG, Woolf CJ, Waxman SG. Two independent mouse lines carrying the Nav1.7-I228M gain-of-function variant display DRG neuron hyperexcitability but a minimal pain phenotype. Pain. 2020 162:1758-1770.
Cheng YC, Snavely A, Barrett LB, Zhang X, Herman C, Frost DJ, Riva P, Tochitsky I, Kawaguchi R, Singh B, Ivanis J, Huebner EA, Arvanites A, Oza V, Davidow L, Maeda R, Sakuma M, Grantham A, Wang Q, Chang AN, Pfaff K, Costigan M, Coppola G, Rubin LL, Schwer B, Alt FW, Woolf CJ. Topoisomerase I inhibition and peripheral nerve injury induce DNA breaks and ATF3-associated axon regeneration in sensory neurons. Cell Rep. 2021 Sep 7;36 :109666
Chiu IM, Heesters BA, Ghasemlou N, Von Hehn CA, Zhao F, Tran J, Wainger B, Strominger A, Muralidharan S, Horswill AR, Wardenburg JB, Hwang SW, Carroll MC, Woolf CJ Bacteria activate sensory neurons that modulate pain and inflammation. Nature. 2013 501:52-57.
Chiu IM, Barrett LB, Williams EK, Strochlic DE, Lee S, Weyer AD, Lou S, Bryman G, Roberson DP, Ghasemlou N, Piccoli C, Ahat E, Wang V, Cobos EJ, Stucky CL, Ma Q, Liberles SD, Woolf CJ. Transcriptional profiling at whole population and single cell levels reveals somatosensory neuron molecular diversity. Elife. 2014 Dec 19;3. doi: 10.7554/eLife.04660
Huang X, Roet KCD, Zhang L, Brault A, Berg AP, Jefferson AB, Klug-McLeod J, Leach KL, Vincent F, Yang H, Coyle AJ, Jones LH, Frost D, Wiskow O, Chen K, Maeda R, Grantham A, Dornon MK, Klim JR, Siekmann MT, Zhao D, Lee S, Eggan K, Woolf CJ. Human amyotrophic lateral sclerosis excitability phenotype screen: Target discovery and validation. Cell Rep. 2021 Jun 8;35(10):109224.
Jayakar S, Shim J, Jo S, Bean BP, Singeç I, Woolf CJ. Developing nociceptor-selective treatments for acute and chronic pain. Sci Transl Med. 2021 Nov 10;13(619): eabj9837
Kiskinis E, Sandoe J, Williams LA, Boulting GL, Moccia R, Wainger BJ, Han S, Peng T, Thams S, Mikkilineni S, Mellin C, Merkle FT, Davis-Dusenbery BN, Ziller M, Oakley D, Ichida J, Dicostanza S, Atwater N, Maeder ML, Goodwin MJ, Nemesh J, Handsaker RE, Paull D, Noggle S, McCarroll SA, Joung JK, Woolf CJ, Brown RH, Eggan K. Pathways Disrupted in Human ALS Motor Neurons Identified through Genetic Correction of Mutant SOD1. Cell Stem Cell. 2014 14:781-795
Latremoliere A, Latini A, Andrews N, Cronin SJ, Fujita M, Gorska K, Hovius R, Romero C, Chuaiphichai S, Painter M, Miracca G, Babaniyi O, Remor AP, Duong K, Riva P, Barrett LB, Ferreirós N, Naylor A, Penninger JM, Tegeder I, Zhong J, Blagg J, Channon KM, Johnsson K, Costigan M, Woolf CJ. Reduction of Neuropathic and Inflammatory Pain through Inhibition of the Tetrahydrobiopterin Pathway. Neuron. 2015 86:1393-406.
Liu,Y, Latremoliere, A, Li, X, Zhang, Z, Chen M, Wang, X, Fanf C, Zhu J, Alexandre C, Gao Z, Chen B, Ding, X, Zhou, J-Y, Zhang Y, Chen C, Wang KH, Woolf CJ, He Z. Touch and tactile neuropathic pain sensitivity are set by corticospinal projections. Nature 2018 561:547-550.
Michoud F, Seehus C, Schönle P, Brun N, Taub D, Zhang Z, Jain A, Furfaro I, Akouissi O, Moon R, Meier P, Galan K, Doyle B, Tetreault M, Talbot S, Browne LE, Huang Q, Woolf CJ, Lacour SP. Epineural optogenetic activation of nociceptors initiates and amplifies inflammation. Nat Biotechnol. 2021 Feb;39:179-185.
Renthal W, Tochitsky I, Yang L, Cheng YC, Li E, Kawaguchi R, Geschwind DH, Woolf CJ. Transcriptional Reprogramming of Distinct Peripheral Sensory Neuron Subtypes after Axonal Injury. Neuron. 2020 108:128-144.
Tegeder I, Costigan M, Griffin RS, Abele A, Belfer I, Schmidt H, Ehnert C, Nejim J, Marian C, Scholz J, Wu T, Allchorne A, Diatchenko L, Binshtok AM, Goldman D, Adolph J, Sama S, Atlas SJ, Carlezon WA, Parsegian A, Lotsch J, Fillingim RB, Maixner W, Geisslinger G, Max MB, Woolf CJ. GTP cyclohydrolase and tetrahydrobiopterin regulate pain sensitivity and persistence. Nature Medicine. 2006 12:1269-1277
Valencia AM, Collings CK, Dao HT, St Pierre R, Cheng YC, Huang J, Sun ZY, Seo HS, Mashtalir N, Comstock DE, Bolonduro O, Vangos NE, Yeoh ZC, Dornon MK, Hermawan C, Barrett L, Dhe-Paganon S, Woolf CJ, Muir TW, Kadoch C. Recurrent SMARCB1 Mutations Reveal a Nucleosome Acidic Patch Interaction Site That Potentiates mSWI/SNF Complex Chromatin Remodeling. Cell. 2019 Nov 27;179(6):1342-1356.
Wainger BJ, Kiskinis E, Mellin C, Wiskow O, Han SS, Sandoe J, Perez NP, Williams LA, Lee S, Boulting G, Berry JD, Brown RH Jr, Cudkowicz ME, Bean BP, Eggan K, Woolf CJ. Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep. 2014 Apr 10;7(1):1-11.
Wainger BJ, Macklin EA, Vucic S, McIlduff CE, Paganoni S, Maragakis NJ, Bedlack R, Goyal NA, Rutkove SB, Lange DJ, Rivner MH, Goutman SA, Ladha SS, Mauricio EA, Baloh RH, Simmons Z, Pothier L, Kassis SB, La T, Hall M, Evora A, Klements D, Hurtado A, Pereira JD, Koh J, Celnik PA, Chaudhry V, Gable K, Juel VC, Phielipp N, Marei A, Rosenquist P, Meehan S, Oskarsson B, Lewis RA, Kaur D, Kiskinis E, Woolf CJ, Eggan K, Weiss MD, Berry JD, David WS, Davila-Perez P, Camprodon JA, Pascual-Leone A, Kiernan MC, Shefner JM, Atassi N, Cudkowicz ME. Effect of Ezogabine on Cortical and Spinal Motor Neuron Excitability in Amyotrophic Lateral Sclerosis: A Randomized Clinical Trial. JAMA Neurol. 2021 78:18